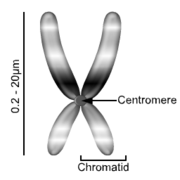
HOME HISTORY COSMOLOGY THEORY DIVINATION GENOME RESOURCE VIDEO THEATRE BIO CONTACT
Contact us at info@lotusspace.com
YIJING HEXAGRAMS AND THE GENETIC CODE:
Mathematical, Structural, and Statistical Analogies; (Schonberger), (Yan)
The biochemical structures of RNA and DNA genetic molecules correspond to the structural components of Yijing hexagrams. Since there is no coincidence from the universal perspective of the Tao, this structural correspondence suggests that there is a mathematical structure inherent in all natural phenomena.
The genetic coding is a vehicle of replication and continuity. The DNA helix is a set of fixed geometrical proportions. Therefore, physical existence is determined by the invisible immaterial world of pure geometrical patterns of wave form.
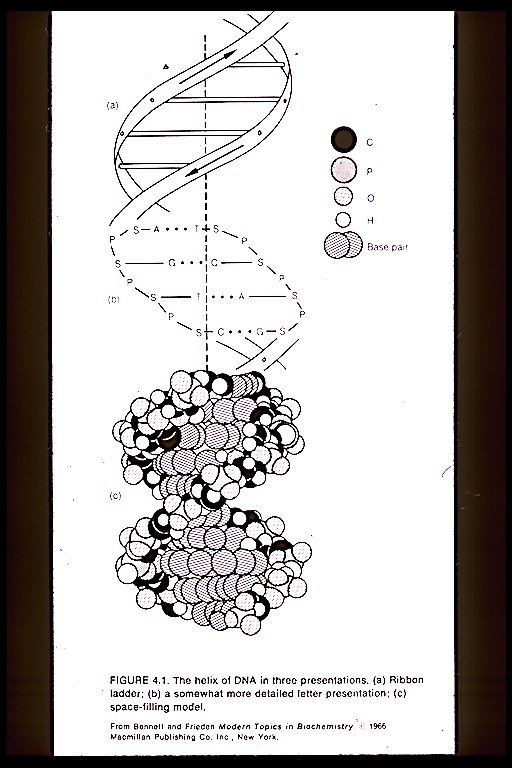
This section examines cell anatomy, biochemistry of the genetic code, Yijing correspondence with the genetic code, genetic structural correspondences, information flows, and pattern correspondences of amino acid number properties with coding properties.
Child = message
Conception = divination
Information
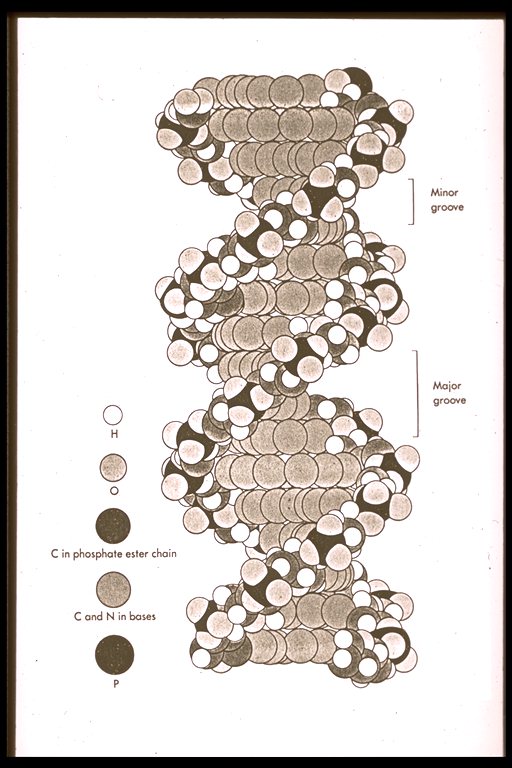
Briefing on Cell Anatomy:
The briefing on cell anatomy shall include cell organelles, membrane proteins, and membrane transportation mechanisms.
Organelles of Cell:
ORGANELLES OF A CELL |
||
Structure |
Composition |
Function |
|
Cell Membrane |
Lipid bilayer containing phospholipids, steroids, and proteins |
Isolation, protection, sensitivity, support: controls entrance/exit of materials |
|
Cytosol |
Fluid component of cytoplasm |
Distributes materials by diffusion |
Nonmembranous Organelles |
||
|
Cytoskeleton: Microtubule, Microfilament |
Proteins organized in fine filaments or slender tubes |
Strength, movement of cellular structures and materials |
|
Microvilli |
Membrane extensions containing microfilaments |
Increase surface area to facilitate absorption of extracellular materials |
|
Cilia |
Membrane extensions containing 9 microtubule doublets + a central pair |
Movement of materials over surface |
|
Centrioles |
Two centrioles, at right angles; each composed of 9 microtubule triplets |
Essential for movement of chromosomes during cell division |
|
Ribosomes |
RNA + proteins; fixed ribosomes bound to endoplasmic reticulum, free ribosomes scattered in cytoplasm |
Protein synthesis |
Membranous Organelles |
||
|
Endoplasmic Reticulum (ER) |
Network of membranous channels extending throughout the cytoplasm |
Synthesis of secretory products; intracellular storage and transport |
|
Rough ER |
Has ribosomes attached to membranes |
Secretory protein synthesis |
|
Smooth ER |
Lacks attached ribosomes |
Lipid and carbohydrate synthesis |
|
Golgi Apparatus |
Stacks of flattened membranes (saccules) containing chambers (cisternae) |
Storage, alteration, and packaging of secretory products and lysosomes |
|
Lysosomes |
Vesicles containing powerful digestive enzymes |
Intracellular removal of damaged organelles or pathogens |
|
Mitochondria |
Double membrane, w/inner folds (cristae) enclosing important metabolic enzymes |
Produce 95% of ATP required by cell |
Nucleus |
Nucleoplasm containing nucleotides, enzymes, and nucleoproteins; surrounded by a double membrane |
Control of metabolism; storage and processing of genetic information; control of protein synthesis |
|
Nucleolus |
Dense region in nucleoplasm containing DNA and RNA |
Site of rRNA synthesis and assembly of ribosomal subunits |
Nuclear Organelles |
||
|
Envelope |
Double membrane containing nucleus and nucleoplasm (nuclear fluid) |
Communicates with cytosol through nuclear pores |
|
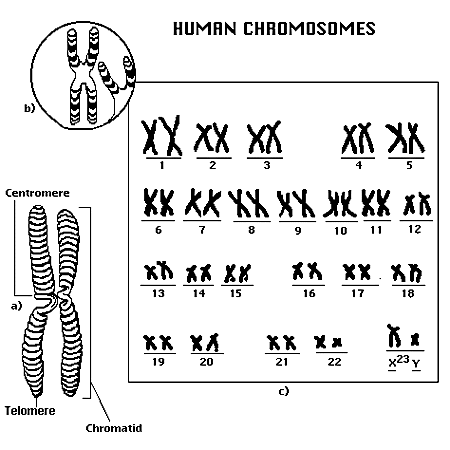
Chromosomes: dividing cells Chromatin: non-dividing cells |
23 pairs per nucleus, containing DNA strands binding histone proteins |
Stores information in DNA for protein synthesis, used for controlling the cell |
Membrane Proteins:
A cell membrane is made up of lipids, proteins and carbohydrates.
|
MEMBRANE PROTEINS |
||
Class |
Function |
Example |
|
Receptor Proteins |
Sensitive to specific extracellular materials that bind to them and trigger a change in cell activity |
Insulin binding to membrane receptors increases the rate of glucose absorption by the cell |
|
Channel Proteins |
Central pore, or channel, permits water and solutes to bypass lipid portion of cell membrane |
Calcium ion movement through channels is involved in muscle contraction and the conduction of nerve impulses |
|
Carrier Proteins |
Bind and transport solutes across the cell membrane; may or may not require energy |
Carriers bring glucose into cytoplasm, and transport sodium, potassium, and calcium ions |
|
Enzymes |
Catalyze reactions in the extracellular fluid or w/in the cell |
Dipeptides are broken down into amino acids by enzymes on the membranes of cells lining the intestines |
|
Anchor Proteins |
Attach the cell membrane to other structures for position stability |
Inside: bound to cytoskeleton; Outside: extracellular proteins of foreign cell |
|
Identifier Proteins |
Identify a cell a self or nonself, normal or abnormal, to the immune system |
Major Histocompatibility Complex (MHC) |
Membrane Transportation Mechanisms:
TRANSPORTATION MECHANISMS ACROSS THE CELL MEMBRANE |
|||
Mechanism |
Process |
Factors Affecting Rate |
Substances Involved |
|
DIFFUSION |
Molecular movement of solutes; direction determined by relative concentrations |
Size gradient, molecular size, charge, lipid solubility |
Small inorganic ions, lipid soluble materials (all cells) |
|
Osmosis |
Movement of water molecules toward solution containing relatively higher solute concentration; requires membrane |
Concentration gradient, opposing osmotic or hydrostatic pressure |
Water only (all cells) |
|
FILTRATION |
Movement of water, usually with solute, by hydrostatic pressure; requires filtration membrane |
Amount of pressure, size of pores in filter |
Water and small ions (blood vessels) |
|
CARRIER MEDIATED TRANSPORT |
|||
|
Facilitated (Passive) Diffusion |
Carrier molecules passively transport solutes down a concentration gradient |
As above, plus availability of carrier protein |
Glucose and amino acids (all cells) |
|
Active Transport |
Carrier molecules actively transport solutes regardless of any concentration gradients |
Availability of carrier, substrate, and ATP |
NA+, K+, Ca2+, Mg2+ (all cells); other solutes by specialized cells |
|
VESICULAR TRANSPORT |
|||
|
Endocytosis: Pinocytosis: cell drinking Phagocytosis: cell eating |
Creation of vesicles containing fluid or solid material |
Stimulus and mechanics incompletely understood; requires ATP |
Fluids, nutrients (all cells); debris, pathogens (specialized cells) |
|
Exocytosis |
Fusion of vesicles containing fluids and or solids with the cell membrane |
Sim to above |
Fluids, debris (all cells) |
This section on the biochemistry of the genetic code defines the genetic code, biopolymer structures, proteins, RNA, DNA, information transfer, transcription, translation, mitochondrial protein synthesis, and viruses.
Genetic Code:
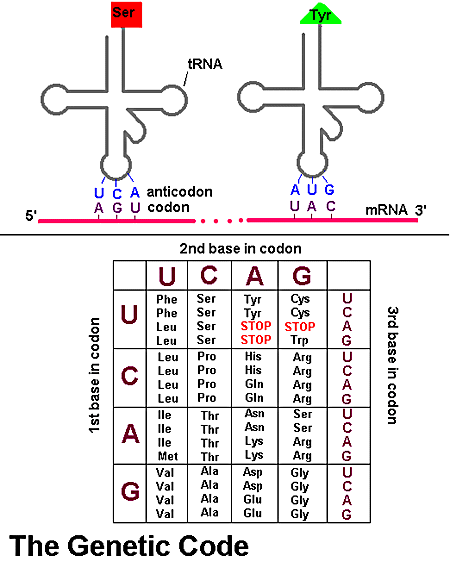
The genetic code, or triplet code, is the information storage system of the cell. The triplet code consists of a sequence of three nucleotide, or nitrogenous, bases which identifies a single amino acid. Each gene consists of all the triplets needed to produce a specific peptide chain.
Biopolymer Structures:
The genetic code, expressed in helical and tertiary structures of protein, is composed of molecules called polymers. Polymers are composed of small structural units, which join in head-to-tail fashion, giving rise to linear polymers. Linear polymers may contain other reactive parts, arms, in addition to heads and tails, resulting in branched polymers. Smaller polymers, oligomers are specified as monomers, dimers, trimers, etc., corresponding to their repeating units. While linear polymers are water-soluble, branched polymers are insoluble in the same solvent, as a gel responsible for blood clotting.
Linear polymers, or macromolecules, more common than branched polymers, are composed of chemical elements: carbon, hydrogen, oxygen, nitrogen, sulfur, and phosphorus.
Proteins:
Proteins are polymers of amino acids. Amino acids have a central carbon, or asymmetric carbon, linked to four different possible groups: head amino group (H-N-H), tail acidic group (-COOH), and side chain groups (H) and (-R). Amino acid differences are distinguished by R group differences. Living systems have 20 different side chains to form 20 different amino acids. This amino acid structure is visualized with the asymmetric carbon in the center of a tetrahedron, with the N, C, R, H groups at the four vertexes. From the perspective of H vertex towards the center, if the other three vertexes, N, C, and R, are arranged clockwise, then the amino acid naturally has a left-handed configuration. All other configurations are right-handed polymers, which can only be synthesized.
Amino acids in a protein molecule join by peptide bonds, which form by eliminating a water molecule (H-O-H), resulting in a dipeptide, or polypeptide bonds.
Proteins biochemical function within the living organism include: assist in the assembling of nucleic acids, transport energy and chemicals, break down large molecules, and synthesize them from smaller constituents. Chemical reactions involving proteins occur in an aqueous environment at a constant body temperature (37°C). Proteins that catalyze (accelerate) reactions are enzymes. Enzyme reactions are very specific to their geometry, like a key and lock. The enzyme shape specifies what reactant (water, amino acid and other proteins) can enter into the active site, to form or degrade of a chemical bond. Enzymic shape and site are the constraints of the template reaction characterizing biochemical activities.
RNA:
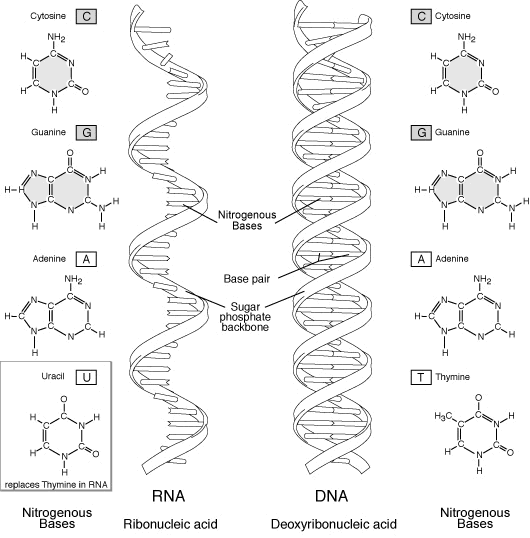
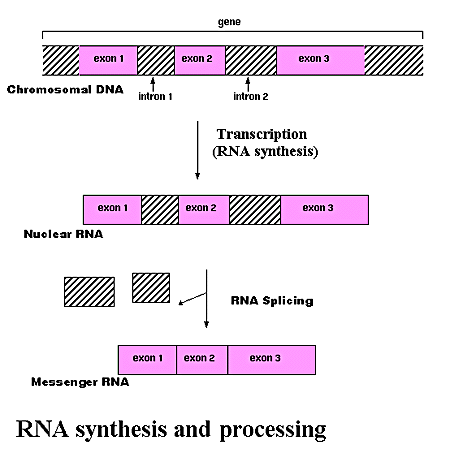
RNA (ribonucleic acid) is a polynucleotide (many short chained units). Each nucleotide consists of a nitrogen base, a five carbon sugar (ribose in RNA, and deoxyribose in DNA), and a phosphate group. The sugar and phosphate group constitutes the backbone of the nucleic acid chain. The four bases of RNA include: adenine (A), cytosine (C), uracil (U), guanine (G). Primordial nucleic acid chains are oligonucleotides, which have exons, protein coding regions, and introns, non-coding regions. Crude RNA, containing exons and introns, eliminates introns by splicing together only the exons, to become a more efficient coding. RNA is more reactive than DNA due to the presence of a hydroxyl (-OH) group attached to a 2 carbon group, as opposed to just a hydrogen atom in DNA.
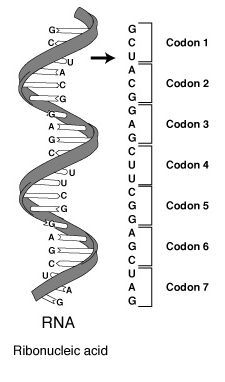
Cells with a nucleus, eukaryotes, are found in multi-celled animals and plants. Cells without a nucleus, prokaryotes, are found in single cell organisms, like bacteria. Both kinds of cells require three types of RNA for transcription and translation: transfer RNA (tRNA), messenger RNA (mRNA), and ribosomal RNA (rRNA), or RNA polymerase. All three are made by the transcription of one DNA double helix. Eukayote cells contain crude RNA with introns and exons, while prokaryote cells contain efficient RNA with only exons.
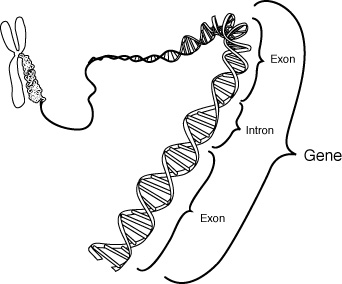
Evolution of RNA and the Genetic Code:
Nucleotides® Oligonucleotides (RNA)® Crude RNA
DNA ¬Crude RNA (exons + introns)® Spliced RNA
¯ Translation ¯
Peptides Protein
DNA:
DNA (deoxyribonucleic acid), the master molecule of life, have four kinds of bases also: adenine (A), cytosine (C), guanine (G), and thymine (T). This molecule exists in a double-stranded helical structure, where the strands are connected by complementary pairings of bases: A to T, and C to G.
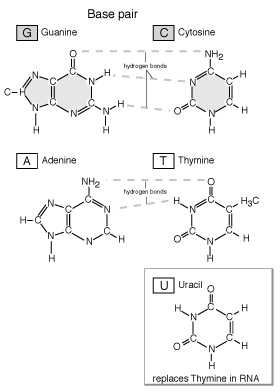
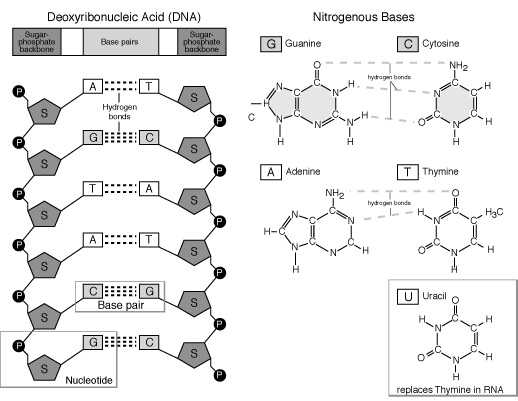
Information Transfer:
Replication ADNA Transcription® RNA Translation® Protein
Transcription:
Definition:
Transcription is the process of forming a strand of mRNA, which carries instructions from the gene in the nucleus to the ribosomes in the cytoplasm.
Steps:
1) Two DNA strands separate, and an enzyme, RNA polymerase, binds to the control segment of the gene.
2) The RNA polymerase moves from one triplet to another along the length of the gene. The RNA polymerase then strings the arriving nucleotides together into a strand of mRNA.
3) Upon reaching the stop signal at the end of the gene, the RNA polymerase and the mRNA strand detach, and the two DNA strands re-associate.
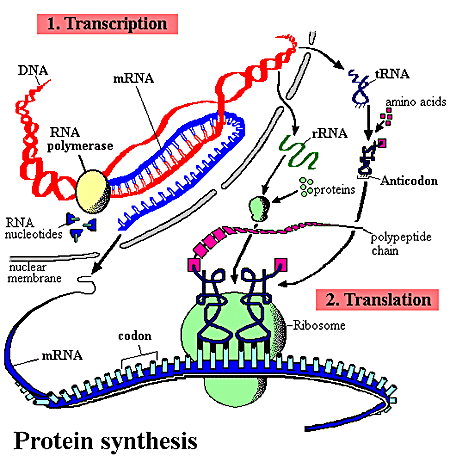
Explanation:
Each DNA strand contains thousands of individual genes. When a gene is activated, an enzyme, RNA polymerase, binds to the initial segments of the gene. This enzyme promotes the synthesis of an mRNA, using nucleotides complementary to those in the gene. RNA polymerase may only attach with RNA bases: A, G, C, or U. The mRNA strand contains a sequence of nitrogen bases complementary to those of the gene. A sequence of three nitrogen bases along the new mRNA strand represents a codon, which complements the corresponding gene triplet. Upon reaching the stop signal, the enzyme and mRNA strand detach, and the complementary DNA strand re-associate. An mRNA strand formed in this way may be altered before it leaves the nucleus.
Translation:
Definition:
During translation a functional polypeptide is constructed using the information from an mRNA strand. Each trio of nitrogen bases along the mRNA strand is a codon. The sequence of codons determines the sequence of amino acids in the polypeptide. Molecules of tRNA bring amino acids to the ribosomes involved in translation.
Steps:
1) A molecule of tRNA brings amino acid (1) to the active binding site at the ribosome. The anticodon of the tRNA must be complementary to the codon on the mRNA strand.
2) As the strand moves across the active site, tRNA-2 arrives with amino acid (2).
3) Enzymes of the ribosome break the linkage between tRNA-1 and amino acid (1) and join amino acids 1 and 2 with a peptide bond. The ribosomes shifts one codon to the right, tRNA-1 departs, without amino acid (1), and tRNA-3 arrives.
4) This process continues until the ribosome reaches the stop codon. The ribosome then breaks the connection between the last tRNA molecule and the peptide chain. The ribosome disengages, leaving the mRNA strand intact.
Explanation:
Translation is initiated when newly synthesized mRNA binds with a ribosome. Molecules of tRNA then deliver amino acids, that the ribosome will assemble into a peptide chain. There are 20 different tRNA for each of the 20 amino acids used in protein synthesis. Each tRNA contains a complementary trio of nitrogen bases, the anticodon, that binds to a specific codon on the mRNA.
Translation begins when the codon of an mRNA strand binds with the first tRNA anticodon. During the consecutive arrival and removal of tRNA, the ribosome enzymes form peptide bonds between amino acids, until the ribosome reaches the stop codon on the mRNA strand. After detachment from the ribosome tRNA can reenter the cytosol, where it can pick up another amino acid and repeat the process. The ribosome then detaches, leaving an intact strand of mRNA and a completed polypeptide.
Translation proceeds as rapidly as metabolism, producing a typical protein (1000 amino acids) in about 20 seconds.
Mitochondrial Protein Synthesis:
In the cytoplasms of eukaryotes are prokaryotes called mitochondria, responsible for energy generation. Prokaryotes use the mitochondrial code, whereas eukaryotes use the nucleic code, or universal genetic code. The mitochondrial code is more symmetrical than the nucleic code with respect to encoded amino acids. Since mitochondria are inherited only from the female parent, and there DNA sequences are short, the DNA sequence variation in mitochondria has significant implications in anthropological evolution.
Degree of Gene Reaction:
Highly Conservative Mitochondrial (prokaryote)® Nuclear (eukaryote)® Highly Innovative Prokaryote
Viruses:
The living cell contains the essential three biopolymers: RNA, DNA, and protein. Viruses may not be called cells for they only carry either DNA or RNA, and are thus classified accordingly. They may be further classified according to the number of nucleic acid strands. Viruses cannot synthesize protein or generate energy. The protein coat of a virus protects it from enzymatic attacks and transports it to susceptible host cells. Virus reproduction occurs within the host cell after invasion.
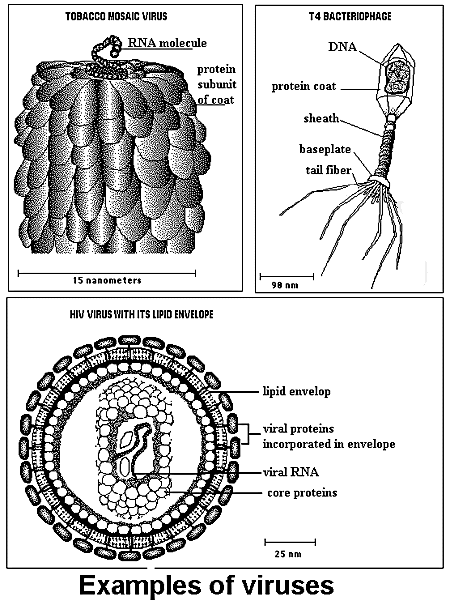
Yijing Genetic Code Mathematical Correspondences:
Mathematical correspondences are examined through number representations, binary system, logarithms information-bearing molecules, and information-bearing sequence constructed with prime numbers.
Number Representations:
1) Odd Numbers (1, 3, 5, 7, 9) = yang = heaven
2) Even Numbers (2, 4, 6, 8, 10) = yin = earth
3) Sum of odd numbers = 25
4) Sum of even numbers = 30
5) Grand Sum = 55 (grand number from which the sum of the ritual numbers must be subtracted to determine the MPML)
Binary System:
The foundation of Yijing mathematics is equal to the binary system of numbers and its use of probabilities.
6) Yin = 0
7) Yang = 1
The Yijing can explain almost anything through the construction of the binary numbers with the power of 2. The binary progression of numbers is seen in the Fu Xi sequence of hexagrams.
Logarithms Information-bearing Molecules:
8) 4 nucleotide bases in DNA and RNA: Information of DNA = log (4) = 2
9) 20 natural amino acid residues in proteins: Information of proteins = log (20) = 4.32
10) Nucleotide bases required to code an amino acid residue: 4.32/2 = 2.16
11) Total number of triplets that can be formed by 4 nucleotide bases: 43 = 4x4x4 = 64
Information-bearing Sequence Constructed with Prime Numbers:
Prime numbers are divisible only by themselves and one. Between 1 and 64 there are 18 primes.
Sequence of Primes between 1-64:
[1] (1) 2 (1) 3 (2) 5 (2) 7 (4) 11 (2) 13 (4) 17 (2) 19 (4) 23 (6) 29 (2) 31 (6) 37 (4) 41 (2) 43 (4) 47 (6) 53 (6) 59 (2) 61 (3) [64]
The differences between primes (numbers in parentheses), show the degeneracy, or numbers of synonymous codons in the genetic code.
DEGENERACY IN THE GENETIC CODEAND DIFFERENCES BETWEEN PRIME NUMBERS IN (1,64) |
|||
|
Differences between Primes in (1,64) |
Number of Synonymous Codons (Degeneracy) |
Amino Acids |
Codons |
|
1 |
1 |
Met |
AUG |
|
1 |
1 |
Trp |
UGG |
|
2 |
2 |
Phe |
UUC, UUU |
|
2 |
2 |
Tyr |
UAC, UAU |
|
2 |
2 |
His |
CAC, CAU |
|
2 |
2 |
Gln |
CAA, CAG |
|
2 |
2 |
Asn |
AAC, AAU |
|
2 |
2 |
Lys |
AAA, AAG |
|
2 |
2 |
Asp |
GAC, GAU |
|
- |
2 |
Glu |
GAA, GAG |
|
- |
2 |
Cys |
UGC, UGU |
|
3 |
3 |
Ile |
AUC, AUU, AUA |
|
- |
3 |
Stop |
UAA, UAG, UGA |
|
4 |
4 |
Val |
GUX |
|
4 |
4 |
Pro |
CCX |
|
4 |
4 |
Thr |
ACX |
|
4 |
4 |
Ala |
GCX |
|
4 |
4 |
Gly |
GGX |
|
6 |
6 |
Leu |
CUX, UUA, UUG |
|
6 |
6 |
Ser |
UCX, AGC, AGU |
|
6 |
6 |
Arg |
CGX, AGA, AGG |
|
6 |
- |
- |
|
|
63 Total |
64 |
21 (including stops) |
|
|
Note: X = A, C, U, or G |
|||
Genetic Structural Correspondences:
Typical structural correspondences between genes and hexagrams are digrams with nucleotide bases, ritual numbers with gross categorical amino acids, and hexagrams with codons.
Types:
4 Digrams » 4 Nucleotide Bases
4 Ritual Numbers » 4 Gross Categories of Amino Acids
64 Hexagrams of the Yijing » 64 Codons of the Genetic Code
Digrams and Nucleotide Bases:
1) Since the purines (nucleotides A and G) are physically larger in molecular dimension, they are assigned “old,” while the pyrimidines (C and U) are smaller and assigned “young.”
2) Pair A-U(T) is equivalent to yin, or even with two hydrogen bonds, while pair G-C is yang, or odd with three hydrogen bonds, being denser.
3) In eukaryote cells, poly-A tails are attached to the mRNA chains, suggesting that A is even, or the receptive (yin; 00) member in the A-G duality. Thus G is odd, or creative (yang; 11).
Ritual Numbers and Amino Acids:
4) A series of A (0) is meaningless unless it is preceded by non-zero numbers. Lysine (AAA), or a series of excess zeros, has a high frequency.
5) Chemically, codon GGG and its anticodon CCC form a total of 9 hydrogen bonds. The codon AAA and its anticodon UUU (TTT) form 6 hydrogen bonds. Among other codon-anticodon pairs, the number of hydrogen bonds is either 6, 7, 8, or 9, the same set of numbers as the ritual numbers of the digram lines. The number of H-bonds in a codon-anticodon pair may be considered the ritual number of that codon. The hexagram corresponding to the codon contains three digrams, each with a ritual number. The sum of these three digram ritual numbers ranges from 18 (for 3 old yins) to 27 (for 3 old yangs).
£ Even and odd number rules: binary code rules
· 3 even numbers = even number
· 3 odd numbers = odd number
· 2 even numbers + 1 odd number = odd number
· 2 odd numbers + 1 even number = even number
Digram Positions Correspondences: Tao of Life
6) In the hexagram, the middle position (human digram) is very significant in fortune determination. Likewise, the second base codon is most important in determining which amino acid is to be coded.
£ Heaven (guiding principle: to live in harmony with nature) = RNA
£ Man (people ceaselessly adapting themselves to the world) = protein
£ Earth (space: environment and evolution) = DNA
DIGRAM-NUCLEOTIDE BASE/RITUAL NUMBERS-AMINO ACID CORRESPONDENCES |
||||
|
Digram |
Old Yin |
Young Yang |
Young Yin |
Old Yang |
|
Binary Number |
00 |
01 |
10 |
11 |
|
Decimal Number |
0 |
1 |
2 |
3 |
|
Nucleotide Base |
Adenine (A) |
Cytosine (C) |
Uracil (U)-RNA Thymine (T)-DNA |
Guanine (G) |
Ritual Number |
6 |
7 |
8 |
9 |
|
Amino Acid Side Chain Polarity |
Large Polar |
Small Non-polar |
Large Non-polar |
Small Polar |
|
Amino Acid Type |
Purine (R) |
Pyrimidine (Y) |
Pyrimidine (Y) |
Purine (R) |
|
Amino Acid Side Chains |
Arginine (Arg) Glutamine (Gln) Glutamate (Glu) Histidine (His) Lysine (Lys) Tryptophan (Trp) Tyrosine (Tyr) |
Isoleucine (Ile) Leucine (Leu) Methionine (Met) Phenylalanine (Phe) Valine (Val) |
Asparagine (Asn) Aspartate (Asp) Glycine (Gly) Serine (Ser) |
Alanine (Ala) Cysteine (Cys) Proline (Pro) Threonine (Thr) |
Hexagrams and Codons:
Correspondence Properties:
7) Symmetry of encoded amino acids:
£ Asymmetrical analogy: nuclear code- probabilities of yarrow stalk method of divination
£ Symmetrical analogy: mitochondrial code- probabilities of coin method of divination
8) Frequency of codon usage: Computation of codon appearance in a particular DNA coding
9) Code development: evolution of the archetypal code into the nuclear and mitochondrial codes
|
EVOLUTION OF THE ARCHETYPAL CODE |
||||
|
Codon Quartets |
Archetypal Code |
Changes in Codon |
Present Code |
|
|
Mitochondrial |
Nuclear |
|||
|
AAX |
Lys |
AAR |
Lys |
Lys |
|
|
|
AAY |
Asn |
Asn |
|
ACX |
Thr |
--- |
Thr |
Thr |
|
AUX |
Ile |
AUA |
Met |
Ile |
|
|
|
AUG |
Met |
Met |
|
|
|
AUY |
Ile |
Ile |
|
AGX |
Ser or Arg |
AGR |
Stop |
Arg |
|
|
|
AGY |
Ser |
Ser |
|
CAX |
His |
CAR |
Gln |
Gln |
|
|
|
CAY |
His |
His |
|
CCX |
Pro |
--- |
Pro |
Pro |
|
CUX |
Leu |
--- |
Leu |
Leu |
|
CGX |
Arg |
--- |
Arg |
Arg |
|
UAX |
Stop |
UAR |
Stop |
Stop |
|
|
|
UAY |
Tyr |
Tyr |
|
UCX |
Ser |
--- |
Ser |
Ser |
|
UUX |
Phe |
UUR |
Ser |
Ser |
|
|
|
UUY |
Phe |
Phe |
|
UGX |
Cys |
UGA |
Trp |
Stop |
|
|
|
UGG |
Trp |
Trp |
|
|
|
UGY |
Cys |
Cys |
|
GAX |
Asp or Glu |
GAR |
Glu |
Glu |
|
|
|
GAY |
Asp |
Asp |
|
GCX |
Ala |
--- |
Ala |
Ala |
|
GUX |
Val |
--- |
Val |
Val |
|
GGX |
Gly |
--- |
Gly |
Gly |
|
Notes: X = A, C, U, or G R = purines (A or G) Y = pyrimidines (C or U) --- = no change |
||||
10) Working principles of Yijing oracle: Yangs and Primes Interpretation
£ Rules for Generating Amino Acids with Prime Numbers (Using the nuclear code):
· The purines, or amino acids created by old yin or old yang, are paired, and the pyrimidines, or amino acids created by young yin or young yang, are paired.
· The prime numbers larger than 2 are also odd numbers, thus creative yang numbers. They correspond to the amino acids created, with the accompanying yin, or even, numbers as their synonymous codons.
£ Rule Exceptions: Chemical Interpretation
· Asymmetric quartets: due to complicated reactions of precursor amino acids
Met (AUX)
Trp (UGX)
· 3 sets of 6-fold degenerate codons
Asp 49-50® Asn (2)-1
Glu 51-48® Gln (19)-16
Phe (41)-42® Tyr 33-34
Ser is coded by UCX’(37) + AGY’(13)
Arg is coded by CGX” + AGR or by CGY’ + (CGR + AGR)’
Leu is coded by UUR’ + CUX or by YUR’ + CUY
· Notes on Primes in Codon Quartets:
· 1 prime per quartet = (‘)
· 2 primes per quartet = (“)
11) Informational Sequences: The gene structure and Yijing divination are different languages using the same set of 64 characters. Consciousness, or information processing, is the dominant task of living nucleic acids within cells.
£ Gene Structure (Life Code): The combination of the four nucleotide bases in a DNA sequence, generates the triplet codon of an amino acid, and conveys a sequential message that determines the structure and function of the protein coded. The genetic code, or primary protein structure, specifies the relationship between nucleotide bases and amino acids. Genes are the blueprint of an individual’s hereditary potential.
£ Yijing Divination (Mind Code): Consciousness plays a significant part in the prediction of a particular time in an individual’s life.
Information Flows: Order 1 (cause)® Chaos® Order 2 (effect)
Both flows have a chaotic, or probabilistic step inserted. The 64 characters lie between chaos and order in both flows.
Genetic Message Coding:
Order 1 Chaos Order 2
Transcription® Translation® low-tech template® High-tech template
(64 Codons) (Enzyme Synthesis)
Divination Process:
Order 1 Chaos Order 2
?® Psychological/Social situation® (64) Hexagrams® Moving lines
Pattern Correspondences of Amino Acid Number Properties and Coding Properties:
The pattern correspondences of amino acid and coding properties follow rules inherently used in amino acid number groups.
Rules:
1) Creative Yang Number Rule: In order for an amino acid to be creative, the amino acid number must be an odd, prime, or both. The first odd number (1) and the only even prime (2) should be used. The stop codons take the number (0).
2) P1 Prime Rule: All P1 primes smaller than 64 are amino acid quartet numbers for 4-fold, or quartet, codons for a given amino acid specified by the first two bases in three codon positions.
£ Explanation: Natural numbers (positive integers)/2 yield remainder 1 for odd numbers or remainder 0 for even numbers.
£ Natural numbers/4 yield quaternary remainders (0, 1, 2, or 3).
£ Odd numbers/4 yield quaternary remainders (1 or 3: P1 or P3).
£ Prime numbers with quaternary remainder 1 are called P1 numbers since they can be expressed as sums of two squares. Each sum pair of squares is unique to a given P1 number. There are 8 P1 numbers smaller than 64.
£ The eight P1 numbers are analogous to the eight synonymous quartets. P1 numbers are each the sum of an odd number square and an even number square:
5 = 12 + 22
13 = 32 + 22
17 = 12 + 42
29 = 52 + 22
37 = 12 + 62
41 = 52 + 42
53 = 72 + 22
61 = 52 + 62
£ Remember decimal number and nucleotide correspondence: 0 = A; 1 = C; 2 = U; 3 = G
3) P3 Prime Rule: Quaternary numbers with remainder 3 cannot be expressed as sums of two squares.
4) 4n Rule: Differences in amino acid numbers between doublets XXY and XXR (purine and pyrimidine differences) are 4n, with n = 0 for Group 1 and 2 Codons. This rule is violated in the presence of amino acid numbers 0 and 2.
Amino Acid Number Groups:
1) Middle-C Codons: use the P1 Prime Rule with equation given below
£ Amino Acid Quartet Number (Q): Q1 = (2a + 1)2 + (2b)2
£ Possible Variables: a = 0, 1, 2, 3; b = 1
2) Other Synonymous Codons: use P1 Prime Rule with equation given below
£ Amino Acids Quartet Number Equation: Q2 = (2a – 1)2 + (2b)2
£ Possible Variables: a = 1, 3; b = 2, 3; Half the total number of codons (32 out of 64) and more than half of the coded amino acids obey this P1 rule.
3) Other Middle-U and Middle-G Codons: uses P1 Prime Rule with two non-prime squares
32 + 42 = 25 for UUX
32 + 62 = 45 for UUX
£ (See ‘Quartet/Doublet Classification and Hexagram-Codon Correspondence’ Table below for other codons.)
4) Middle-A Codons: Quartets of P3 Prime Rule in the general form 4n + 3
£ Early (evolutionary precursors) Amino Acid Doublet Numbers (D): D1 = 4(2a + 1) + 3
£ Later (chemical conversions) Amino Acids Doublet Numbers: D2 = 8(2a + 1) + 3
£ (See ‘Quartet/Doublet Classification and Hexagram-Codon Correspondence’ Table below for codons.)
5) Quartets AUX and CAX: The remaining 5 numbers, 1 and primes 2, 3, 19, 47, can be specified with the unassigned amino acids coded by AUX and CAX. 1 is the only number that obeys the 4n Rule with its pyrimidine counterpart 45 (Cys).
£ Most Complex Amino Acid:
· Trp = 1
· UGR (Trp) = 1 (mitochondrial code)
· UGG (Trp) = 1, UGA (Stop) = 0 (universal code)
£ Kinetic Codons: 3 and 19 can be expressed 8b + 3 with b = 0 and b = 2, respectively. Factor 8 suggests that later amino acids are being used.
· AUR (Met) = 3 (mitochondrial); initiation codon
· CAR (Gln) = 19
· CAY (His) = 47, CAR (Gln) = 19
· AUY (Ile) = 2, AUR (Met) = 3 (mitochondrial)
· AUY (Ile) = 2, AUA (Ile) = 2 (universal) and AUG (Met) = 3 (universal)
|
Complete List of Amino Acids Numbers |
|
0 (Stop) 1 (Trp) 2 (Ile) 3 (Met) 5 (Thr) 7 (Lys) 11 (Asn) 13 (Pro) 17 (Leu) 15 (Gln) 29 (Ser) 31 (Asp) 37 (Arg) 41 (Val) 43 (Tyr) 47 (His) 53 (Ala) 59 (Glu) 61 (Gly) 25 (Phe) 45 (Cys) |
|
Note: underlined numbers are non-primes. |
|
SIX CORRESPONDENCES IN PROPERTIES |
|
|
Number Properties |
Coding Properties |
|
0 |
Stop Signal |
|
P1 numbers |
Synonymous Quartets |
|
Non-prime 2 square sums |
Early precursor amino acids |
|
P3 numbers |
Early/late amino acids |
|
4n rule |
Third position R/Y |
|
Quaternary units |
Trp and kinetic codons |
|
QUARTET/DOUBLET CLASSIFICATION AND HEXAGRAM-CODON CORRESPONDENCE (In Fu Hsi Sequence) |
|||||
|
General Hexagram Quartet Theme |
Amino Acid Quartet/Doublet (Number and Group): Side Chain Attributes and Function |
||||
|
Hexagram: No. in King Wen Sequence |
Triplet Codon |
Encoded Amino Acid w/Amino Acid No. and Group |
Binary No. w/ Corresponding Decimal No. |
3 Ritual Nos. of Digrams |
Total H-Bonds per Codon-Anticodon Pair |
|
|
AAX (G4): Early AAR = 7; Late AAY = 11 |
||||
|
(2) Kun |
AAA |
Lys (7) |
000000 (0) |
666 |
6 |
|
(23) Bo |
AAC |
Asn (11) |
000001 (1) |
667 |
7 |
|
(8) Bi |
AAU |
Asn (11) |
000010 (2’) |
668 |
6 |
|
(20) Guan |
AAG |
Lys (7) |
000011 (3’) |
669 |
7 |
|
Progress |
ACX = 5 (G1): small non-polar |
||||
|
(16) Yu |
ACA |
Thr (5) |
000100 (4) |
676 |
7 |
|
(35) Jin |
ACC |
Thr (5) |
000101 (5’) |
677 |
8 |
|
(45) Zui |
ACU |
Thr (5) |
000110 (6) |
678 |
7 |
|
(12) Bi |
ACG |
Thr (5) |
000111 (7’) |
679 |
8 |
|
|
AUX* (G5): AUY/AUA = 2 (nuclear); AUG = 3 (mitochondrial) |
||||
|
(15) Qian |
AUA |
Ile (2) |
001000 (8) |
686 |
6 |
|
(52) Gen |
AUC |
Ile (2) |
001001 (9) |
687 |
7 |
|
(39) Jian |
AUU |
Ile (2) |
001010 (10) |
688 |
6 |
|
(53) Jian |
AUG |
Met* (3) |
001011 (11’) |
689 |
7 |
|
|
AGX (G3): AGY = 29; AGR = 37 (nuclear)/ (Stop) = 0 (mitochondrial) |
||||
|
(62) Xiaoguo |
AGA |
Arg (37) |
001100 (12) |
696 |
7 |
|
(56) Lu |
AGC |
Ser (29) |
001101 (13’) |
697 |
8 |
|
(31) Xian |
AGU |
Ser (29) |
001110 (14) |
698 |
7 |
|
(33) Dun |
AGG |
Arg (37) |
001111 (15) |
699 |
8 |
|
|
CAX (G5): CAY = 47; CAR = 19 |
||||
|
(7) Shi |
CAA |
Gln (19) |
010000 (16) |
766 |
7 |
|
(4) Meng |
CAC |
His (47) |
010001 (17’) |
767 |
8 |
|
(29) Kan |
CAU |
His (47) |
010010 (18) |
768 |
7 |
|
(59) Huan |
CAG |
Gln (19) |
010011 (19’) |
769 |
8 |
|
Difficulty |
CCX = 13 (G1): disrupts intra-molecular H-bonds in proteins |
||||
|
(40) Xie |
CCA |
Pro (13) |
010100 (20) |
776 |
8 |
|
(64) Weiji |
CCC |
Pro (13) |
010101 (21) |
777 |
9 |
|
(47) Kun |
CCU |
Pro (13) |
010110 (22) |
778 |
8 |
|
(6) Song |
CCG |
Pro (13) |
010111 (23’) |
779 |
9 |
|
Cooperation |
CUX = 17 (G2): large non-polar |
||||
|
(46) Sheng |
CUA |
Leu (17) |
011000 (24) |
786 |
7 |
|
(18) Gu |
CUC |
Leu (17) |
011001 (25) |
787 |
8 |
|
(48) Jing |
CUU |
Leu (17) |
011010 (26) |
788 |
7 |
|
(57) Sun |
CUG |
Leu (17) |
011011 (27) |
789 |
8 |
|
Duration |
CGX = 37 (G2): large polar (strong alkaline) |
||||
|
(32) Heng |
CGA |
Arg (37) |
011100 (28) |
796 |
8 |
|
(50) Ding |
CGC |
Arg (37) |
011101 (29’) |
797 |
9 |
|
(28) Daguo |
CGU |
Arg (37) |
011110 (30) |
798 |
8 |
|
(44) Gou |
CGG |
Arg (37) |
011111 (31’) |
799 |
9 |
|
|
UAX (G4): UAR = 0 [23]; UAY = 43 |
||||
|
(24) Fu |
UAA |
Stop (0) [23] |
100000 (32) |
866 |
6 |
|
(27) Yi |
UAC |
Tyr (43) |
100001 (33) |
867 |
7 |
|
(3) Jun |
UAU |
Tyr (43) |
100010 (34) |
868 |
6 |
|
(42) Yi |
UAG |
Stop (0) [23] |
100011 (35) |
869 |
7 |
|
Leadership |
UCX = 29 (G1): small polar |
||||
|
(51) Zhen |
UCA |
Ser (29) |
100100 (36) |
876 |
7 |
|
(21) Shihe |
UCC |
Ser (29) |
100101 (37’) |
877 |
8 |
|
(17) Sui |
UCU |
Ser (29) |
100110 (38) |
878 |
7 |
|
(25) Wuwang |
UCG |
Ser (29) |
100111 (39) |
879 |
8 |
|
|
UUX (G3): UUY = 25; UUR = 17 |
||||
|
(36) Mingyi |
UUA |
Leu (17) |
101000 (40) |
886 |
6 |
|
(22) Bi |
UUC |
Phe (25) |
101001 (41’) |
887 |
7 |
|
(63) JiJi |
UUU |
Phe (25) |
101010 (42) |
888 |
6 |
|
(37) Jiaren |
UUG |
Leu (17) |
101011 (43’) |
889 |
7 |
|
|
UGX*: UGY = 45; UGA = 0 (nuclear) (G3); UGR/UGG = 1 (mitochondrial) (G5) |
||||
|
(55) Feng |
UGA |
Stop (0) |
101100 (44) |
896 |
7 |
|
(30) Li |
UGC |
Cys (45) |
101101 (45) |
897 |
8 |
|
(49) Ge |
UGU |
Cys (45) |
101110 (46) |
898 |
7 |
|
(13) Tongren |
UGG |
Trp* (1) |
101111 (47’) |
899 |
8 |
|
|
GAX (G4): GAY = 31; GAR = 59 |
||||
|
(19) Lin |
GAA |
Glu (59) |
110000 (48) |
966 |
7 |
|
(41) Sun |
GAC |
Asp (31) |
110001 (49) |
967 |
8 |
|
(60) Jie |
GAU |
Asp (31) |
110010 (50) |
968 |
7 |
|
(61) Zhongfu |
GAG |
Glu (59) |
110011 (51) |
969 |
8 |
|
Gentility |
GCX = 53 (G1): small non-polar |
||||
|
(54) Guimei |
GCA |
Ala (53) |
110100 (52) |
976 |
8 |
|
(38) Kui |
GCC |
Ala (53) |
110101 (53’) |
977 |
9 |
|
(58) Dui |
GCU |
Ala (53) |
110110 (54) |
978 |
8 |
|
(10) Lu |
GCG |
Ala (53) |
110111 (55) |
979 |
9 |
|
Abundance |
GUX = 41 (G2): large non-polar |
||||
|
(11) Tai |
GUA |
Val (41) |
111000 (56) |
986 |
7 |
|
(26) Dachu |
GUC |
Val (41) |
111001 (57) |
987 |
8 |
|
(5) Xu |
GUU |
Val (41) |
111010 (58) |
988 |
7 |
|
(9) Xiaochu |
GUG |
Val (41) |
111011 (59’) |
989 |
8 |
|
Firmness |
GGX = 61 (G2): smallest (H-atom) |
||||
|
(34) Dazhuang |
GGA |
Gly (61) |
111100 (60) |
996 |
8 |
|
(14) Dayou |
GGC |
Gly (61) |
111101 (61’) |
997 |
9 |
|
(43) Guai |
GGU |
Gly (61) |
111110 (62) |
998 |
8 |
|
(1) Qian |
GGG |
Gly (61) |
111111 (63) |
999 |
9 |
|
Notes: (‘) = prime number (*) = Asymmetric quartets Underlined number = non-prime amino acid numbers Number in [ ] = amino acid number that was over ridden by stop codon number (0) (G#) = Amino Acid Group Number Outer Triplet Codons of Quartet Group (R) = purines (A or G); old yin paired with old yang, respectively Inner Triplet Codons of Quartet Group (Y) = pyrimidines (C or U); young yang paired with young yin, respectively |
|||||